Chapter 4. DNA Replication
DNA and Chromosomes
Apart from reproductive cells (gametes), each cell of the human body contains 23 pairs of chromosomes, each a packet of compressed and entwined DNA. Every strand of the DNA is a huge natural polymer of repeating nucleotide units, each of which comprises a phosphate group, a sugar (deoxyribose), and a base (either adenine, thymine, cytosine, or guanine). Every strand thus embodies a code of four characters (A’s, T’s, C’s, and G’s), the recipe for the machinery of human life.
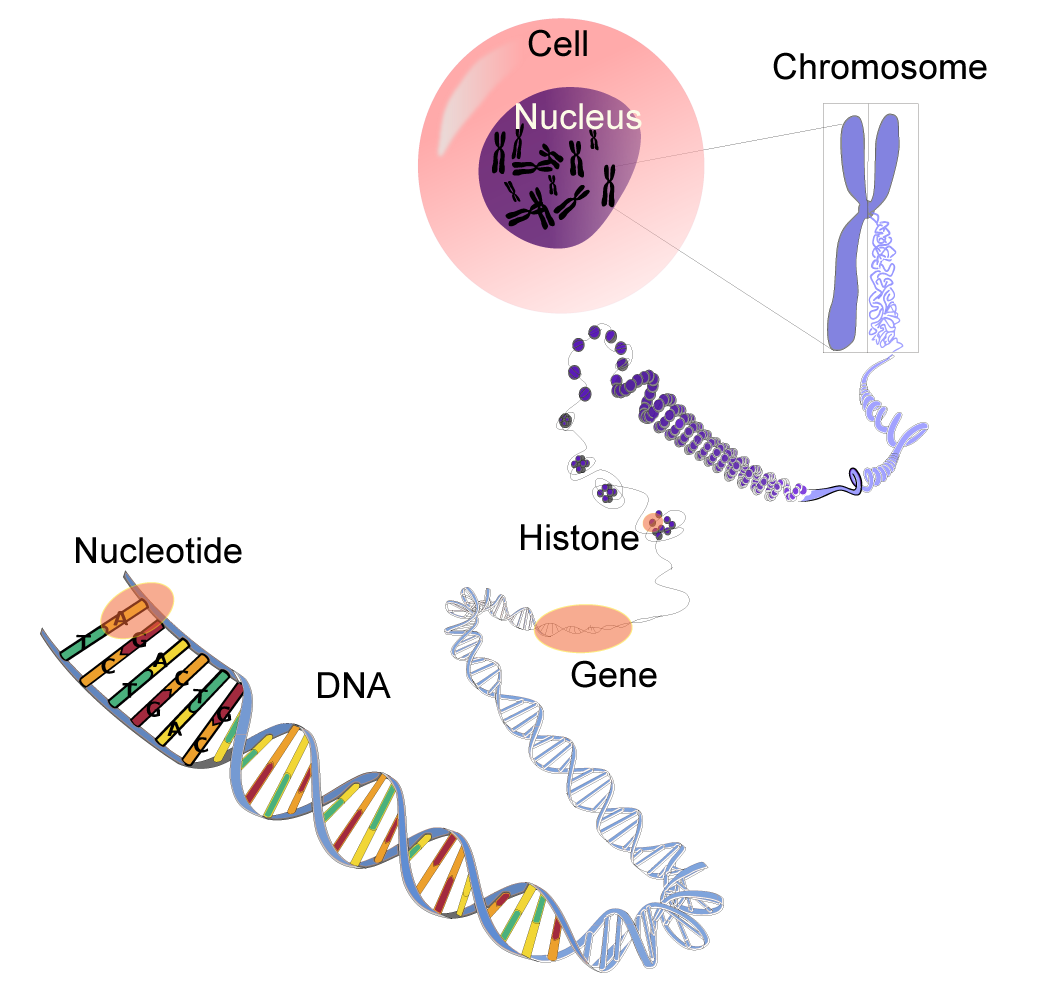
In its normal state, DNA takes the form of a highly regular double-stranded helix, the strands of which are linked by hydrogen bonds between adenine and thymine (A‹T) and between cytosine and guanine (C‹G). Each such linkage is said to constitute
a base pair; some three billion base pairs constitute the human genome. It
is the specificity of these base-pair linkages that underlies the mechanism of DNA replication illustrated in this chapter. Each strand of the double helix serves as a template for the synthesis of a new strand, the nucleotide sequence of which is strictly determined. Replication thus produces twin daughter helices, each an exact replica of its sole parent.
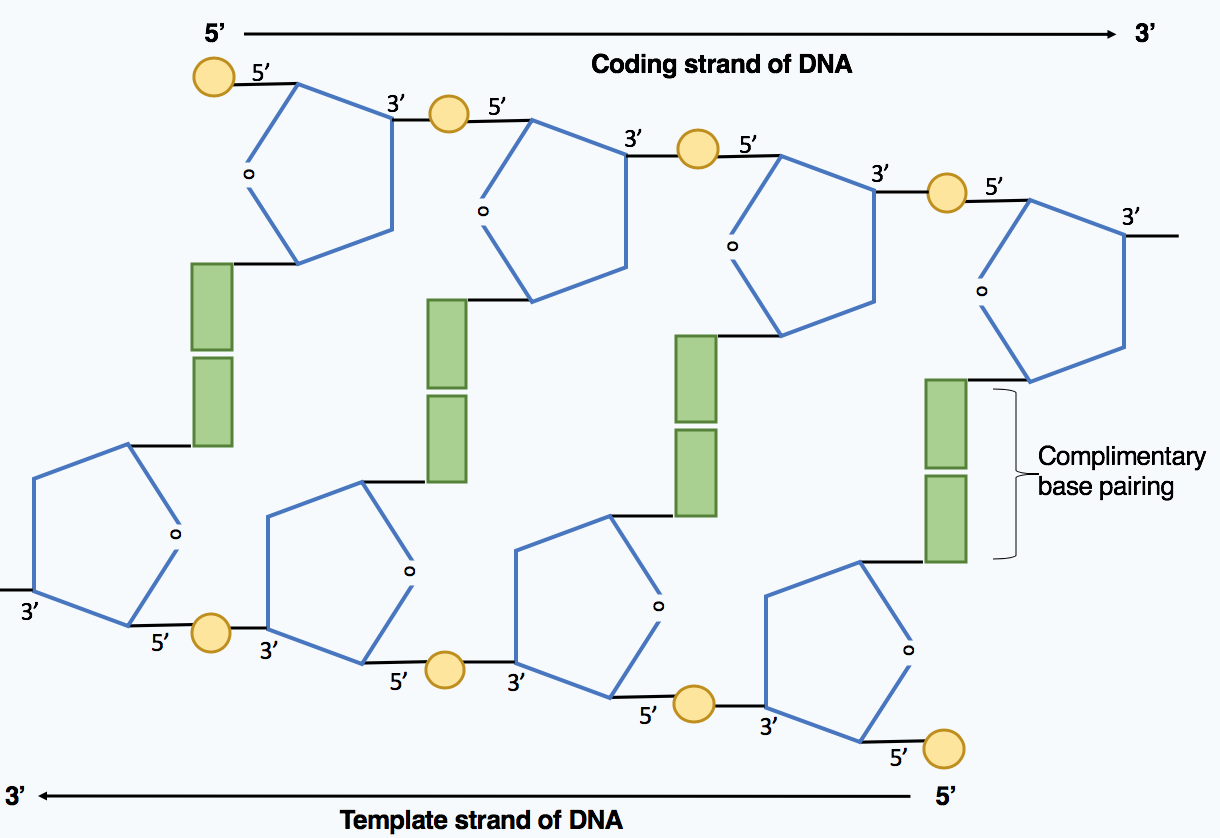
DNA directionality
DNA strands have directionality, and the different ends of a single strand are called the “3′ (three-prime) end” and the “5′ (five-prime) end”. These terms refer to the carbon atom in deoxyribose to which the next phosphate in the chain attaches. In addi- tion to being complementary, the two strands of DNA are antiparallel: They are orientated in opposite directions. This directionality has consequences in DNA synthesis, because DNA polymerase can synthesize DNA in only one direction by adding nucleotides to the 3′ end of a DNA strand.
DNA replication
DNA replication begins at a fixed origin of replication (ori) and usually involves both DNA strands at the same time. The replication is initiated by addition of an RNA primer by the enzyme primase.
In the newly forming nucleic acid strand, nucleotides are added one at a time to the 3’-OH (the 3rd carbon of deoxyribose) end of the growing chain (in 5’ to 3’ direction) by the enzyme DNA polymerase.
The fixed position of the ori sites and the 5’-3’ direction of DNA synthesis makes it possible for only one out of two new DNA strands to be synthesized continuously. This strand is called the leading strand. The other strand (the lagging strand) is synthe- sized in pieces, called the Okazaki fragments.
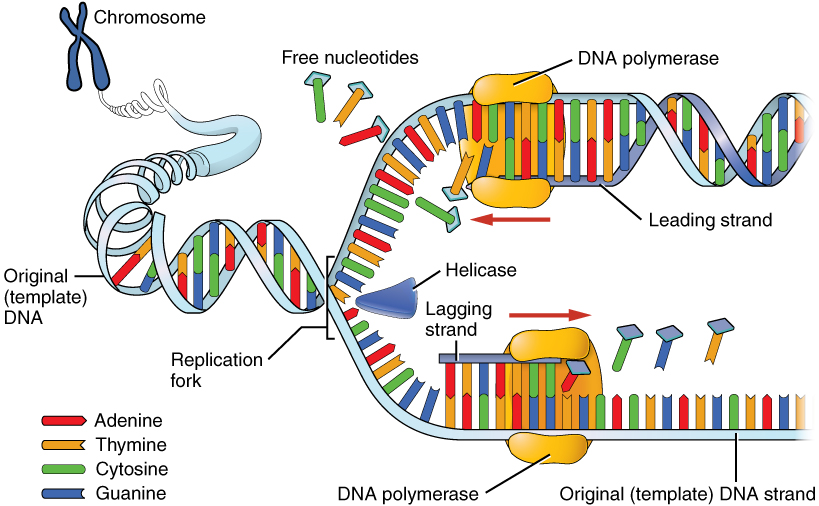
Because DNA replication does not begin at either end of the DNA strand, but starts in the center, and considering that all DNA polymerases that have been discovered synthesize a new DNA strand in the 5′ to 3′ direction, the leading and the lagging strands on the DNA molecule are replicated differently.
On the leading strand, DNA polymerase can make a complementary DNA strand without any difficulty because it goes from 5′ to 3′. However, there is a problem going in the other direction on the lagging strand. To counter this, short sequences of RNA acting as primers attach to the lagging strand a short distance ahead of where the initiation site was. The DNA polymerase can start replication at that point and go to the end of the initiation site. This causes the formation of Okazaki fragments. More RNA primers attach further on the DNA strand and DNA polymerase comes along and continues to make a new DNA strand.
Replication of the ends of DNA molecules
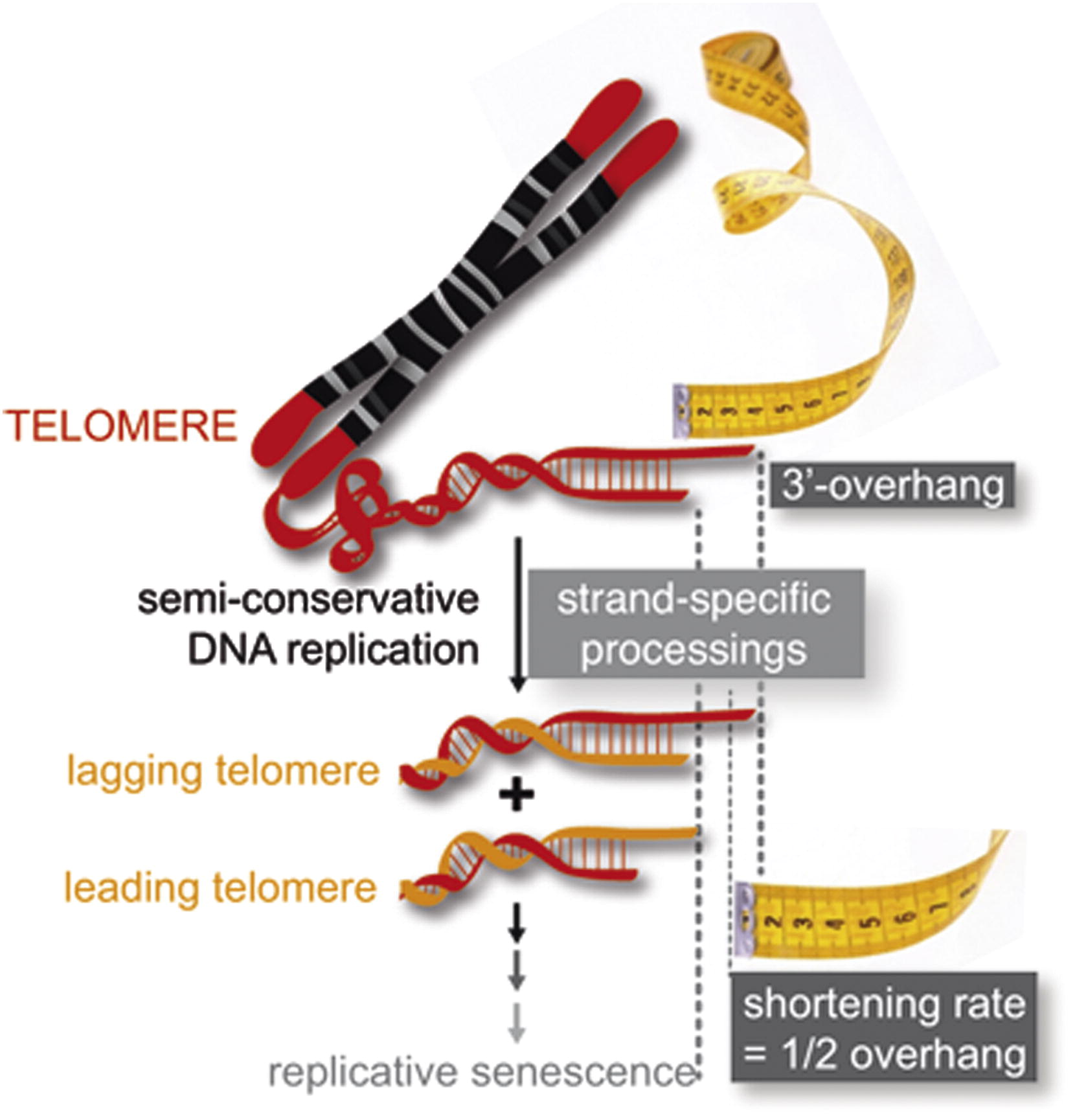
Eventually, the last RNA primer attaches at the end of the lag- ging strand, and DNA polymerase, RNA nuclease (to remove RNA nucleotides of the primer), and DNA ligase come along to convert the RNA (of the primers) to DNA and to seal the gaps in between the Okazaki fragments. But, in order to change RNA to DNA, there must be another DNA strand in front of the RNA primer. This happens at all the sites of the lagging strand, but it does not happen at the 5” end of either newly formed DNA strands where the last (for the lagging strand) or first (for the leading strand) RNA primer is attached. Ultimately, that RNA is destroyed by enzymes that degrade any RNA left on the DNA.
Thus, a section of the telomere is lost during each cycle of replication at the 5′ end of both the leading and the lagging strands. As a result, repeated rounds of replication produce shorter and shorter DNA molecules in somatic cells.
Telomeres
To prevent damage to DNA ends during repeated rounds of replication, eukaryotic chromosomes are capped with repeated DNA sequences called telomeres. In all vertebrates, the sequence is TTAGGG. The repetitive sequences that compose telomeres do not carry any essential genetic information and, thus, their loss during
replication does not result in functional problems for the chromosome. But, with enough rounds of replication, the repetitive ends of chromosomes do “wear out.” When telomeres become too short, a cell can seize any subsequent cell division
and enter into a state of “replicative senescence.”
In reproductive cells, embryonic stem cells and cancer cells, telomere sequences are extended by the enzyme telomerase. Telomerase belongs to a class of replicative enzymes called reverse transcriptases, which use RNA molecule as a template to synthesize a complementary DNA strand. Telomerase adds telomeric sequence repeats (TTAGGG) to the terminal 3′ ends of the template DNA strands anDNA strands and DNA polymerase then extends the telomeric 5’ ends of the newly synthesized strands using the RNA oligonucleotide carried by telomerase as a primer. The existence of a compensatory shortening of telomere (telomerase) mechanism was first predicted by a Russian biologist Alexey Olovnikov in 1973, who also suggested the telomere hypothesis of aging and the telomere’s connections to cancer. Telomerase was discovered by Carol W. Greider and Elizabeth Blackburn in 1984 in the ciliate Tetrahymena. Together with Jack W. Szostak, Greider and Blackburn were awarded the 2009 Nobel Prize in Physiology or Medicine for their discovery.
While the original discoveries of telomerase and its role have concluded that there is little, if any, telomerase activity in somatic cells, this understanding has been challenged in recent years. A NASA study of twin astronauts (the preliminary findings of which were announced in January 2018) revealed that telomeres do get significantly extended during a prolonged space flight. Remarkably, it appears that the majority of those telomeres shorten within a couple of days of the return to Earth.
A recent study determined that the level of telomerase activity is dependent on concentrations of deoxyguanosine diphosphate (dGDP). Guanine happens to be the first nucleotide added by telomerase to the terminal 3’ end of the template DNA strand. A substitution in the RNA template of telomerase leading to the switch in first nucleotide added from G to A leads to a shift in telomerase repeat addition activity stimulation from being dGDP-dependent to being dATP-dependent, leading to a dramatic increase in telomerase activity in somatic cells (Chen et al., 2018).
Role of telomeres and telomerase
Cancer cells are able to escape cellular senescence by rebuilding their telomeres after each replication round. This is possible because cancer cells can reactivate their telomerase, which is normally inactive in adult somatic cells. Several mechanisms of telomerase activation in cancer cells have been proposed. On of these mechanisms is a mutation in the promoter region of the hTERT gene on chromosome 5 that encodes telomerase in humans. When mutated, the hTERT promoter is able to initiate the transcription of this somatically silent gene, leading to the production of functional telomerase.
Telomeres are also involved in tumor initiation and progression. Longer telomeres seem to offset the progression of carcinomas.
Telomeres are involved in aging. Shortening of telomeres shuts down mitosis and leads to progressive “wearing out” of actively dividing tissues. Telomere length could be inherited, contributing to the heritability of longevity. At the same time, longer telomeres encourage malignant proliferation.
Telomeres also participate in the regulation of meiosis. Abnormal telomere extension seems to disrupt meiotic division.
Telomeres are involved in the maintenance of bone marrow function. Telomerase function appears to help maintain and even accelerate osteogenesis in human bone marrow cells.
Key Takeaways
- DNA in human somatic cells undergoes semi-conservative replication before a cell division.
- Telomeric sequences at the end of chromosomes prevent functional DNA from degradation.
A reverse transcriptase (RT) is an enzyme used to generate complementary DNA (cDNA) from an RNA template, a process called reverse transcription. Reverse transcriptases are utilized by viruses such as HIV and hepatitis B to replicate their genomes. Reverse transcriptase is also used by mobile genetic elements to proliferate within the genome, and by eukaryotic cells to extend the telomeres at the ends of their linear chromosomes.
