Chapter 2. The Rules of Genetic Inheritance

The Relationship Between Genotype and Phenotype
This chapter provides an overview of the principles of classical (Mendelian) genetics. The relationship between the physical appearance (phenotype) of a living being and the genetic determinants (factors, as they used to be called) was first described by a Czech monk Gregor Mendel in the 1860s.
- Dominant trait/allele
- Recessive trait/allele
- Homozygous/Heterozygous
- Phenotype/Genotype
Monohybrid cross (follows one character)/Dihybrid cross (two characters are being followed)
Classical (Mendelian) transmission genetics assumes a binary relationship between a character and a gene that encodes that character. Such as, in the classical transmission genetics there are only two versions of the gene dominant of recessive, which controls the phenotype in either expressing the character or not. Correspondently, classical transmission genetics assumes that discrete phenotypes are produces based on the predictable expression of contributing alleles. In this relationship, each allele is either dominant or recessive and it determines one of the alternative phenotypes (white/red, smooth/wrinkled etc.). A homozygous genotype produces the phenotype determined by the corresponding allele, while a heterozygous genotype produces the phenotype determined by the dominant allele.
The OCA2 Gene And The Wild Type Paradigm
Traditionally, the most common phenotype observed in natural populations, and hence its underlaying genotype, was referred to as the wild type. However, when it comes to humans, the designation of a “natural population” and, consequently, the wild type, is problematic. Another way to think about the wild type is that to have it determine the original state of the phenotype (and the corresponding genotype).
To use this designation objectively, we need to have a good idea of the origin and distribution of the original state type, which is aided by the knowledge of the ultimate origin of a species.
When it comes to humans, there is ample scientific evidence that the cradle of the Homo sapiens species is in Africa.
Thus, the most common phenotypes observed in African and African-origin populations would be the most likely candidates for the original state types.
In humans, brown eye color is considered to be the original state type color of the eyes. About three-fourths of eye color variation can be explained by genetic changes in and around the OCA2 gene located on chromosome 15. While there is a wide variability in eye color, colors other than brown only exist among individuals of European descent. African and Asian populations are typically brown-eyed. In 2008 a team of researchers studying the OCA2 gene published results demonstrating that the allele associated with blue eyes oc- curred only within the last 6,000 – 10,000 years within the Euro- pean population (Eiberg et al., 2008). Studies of DNA extracted from ancient human remains confirmed the timing of the blue- color gene. Such as, the remains of a 7,000 year-old Mesolithic hunter gatherer from Spain revealed that he had blue eyes, along with dark hair and skin pigmentation. also suggesting that the light skin pigmentation arose in Europe after the blue eye color mutation came about (Olalde et al., 2014).

Phenotypes & genotypes: the relationship is rarely simple
In some cases, genetics may be difficult to interpret because of the complexity of how a genotype manifests itself as a phenotype.
Factors that affect the Mendelian phenotype/genotype relationship:
Dominance relationships:
- Overdominance (otherwise known as heterosis, hybrid vigor or heterozygote advantage)
- Incomplete dominance
- Codominance
- Multiple alleles
- Epistasis / gene interaction
- Penetrance
- Expressivity
- Mosaicism
- Imprinting
Levels Of Dominance
Overdominance
Overdominance is a condition in genetics where the phenotype of the heterozygote lies outside the phenotypical range of both homozygote parents. Overdominance can also be described as heterozygote advantage, wherein heterozygous individuals have a higher fitness than homozygous individuals. An example in humans is sickle cell anemia.
Sickle cell anemia is a recessive disorder characterized by abnormal (sickle-like) shape of the red blood cells. This change in shape leads to a reduced capacity of erythrocytes to carry oxygen; which leads to a variety of health complications, including a lower life expectancy. The condition is a result of a mutation in the hemoglobin gene. In people heterozygous for the sickle cell allele, the health problems are minor, since the normal allele is able to produce normal hemoglobin.
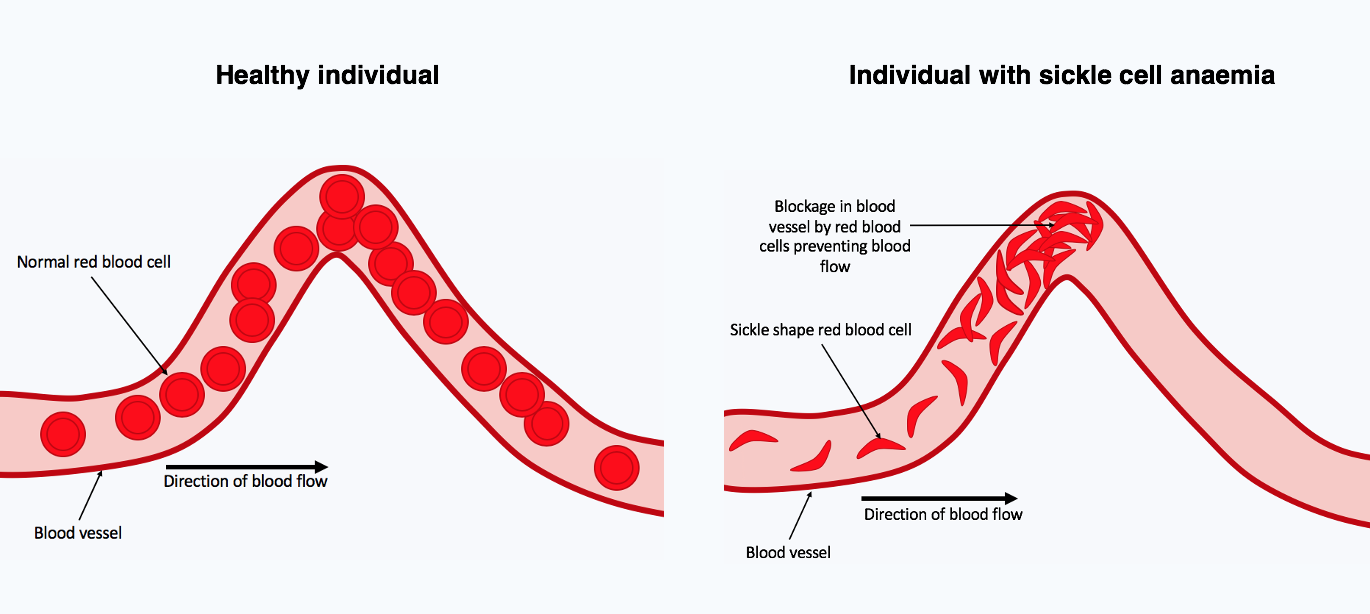
However, this allele also yields some resistance to malaria. Therefore, in regions where malaria exerts or has exerted a strong selective pressure, the sickle cell allele has been selected for its conferred partial resistance to the disease. Since the gene is incompletely recessive, carriers can produce a few sickled red blood cells, not enough to cause symptoms, but enough to give resistance to malaria.
The sickle cell allele seems to have originated independently in at least five times in different geographic locations, such as Atlantic West Africa (the Senegal haplotype), Central West Africa (Benin haplotype), Cameroon (Cameroon haplotype), as well as India (the Adavasi haplotype) and eastern Saudi Arabia.
Incomplete dominance
In classical genetics, one allele is usually considered as completely dominant, one as completely recessive. Some mutations (amorphs or nulls) are completely recessive, but in many cases they have some residual activity (hypomorph or leaky) – incom- plete dominance.
In situations of incomplete dominance, the heterozygote (Aa) has an intermediate phenotype between both homozygotes. One of the most well-studied examples of incomplete dominance in humans occurs in the genes for curly hair. Inheriting a gene for curly hair from one parent and a gene for straight hair from the other parent will give a hair texture that is a blend of the two, wavy hair.

Codominance (additivity)
Codominance occurs when both alleles are separately expressed in the phenotype. In codominance, the heterozygote exhibit the phenotypes of both homozygotes.
A1A2 expresses both the A1 and A2 phenotypes (human MN, ABO blood groups).
Multiple alleles of a single gene
In a population of individuals, a given gene may have several alleles (for example, one wild type and the rest mutant). Some traits are determined by more than one variation of either allele.
Example: There are 60+ different mutations in the CFTR gene that could lead to cystic fibrosis of various degrees of severity (expressivity), depending on a particular mutation.
Although a gene may be represented by multiple alleles in a population, a single individual will still carry only two of these alleles, one on each homologous chromosome.
Example of multiple alleles and co-dominance: ABO blood groups
Antigens are surface markers on the cell membrane that can stimulate an immune response. Antigens can be proteins, carbohydrates, or a combination of the two types of macromolecules (known as glycoproteins).
The red blood cell antigens of the ABO system are carbohydrates. ABO antigens are produced by a series of enzymatic reactions (additions of monosaccharide units) performed by glycosil transferases. The A and B antigens are synthesized from a precursor oligosaccharide (H antigen). The H antigen itself does not stimulate an immune response.
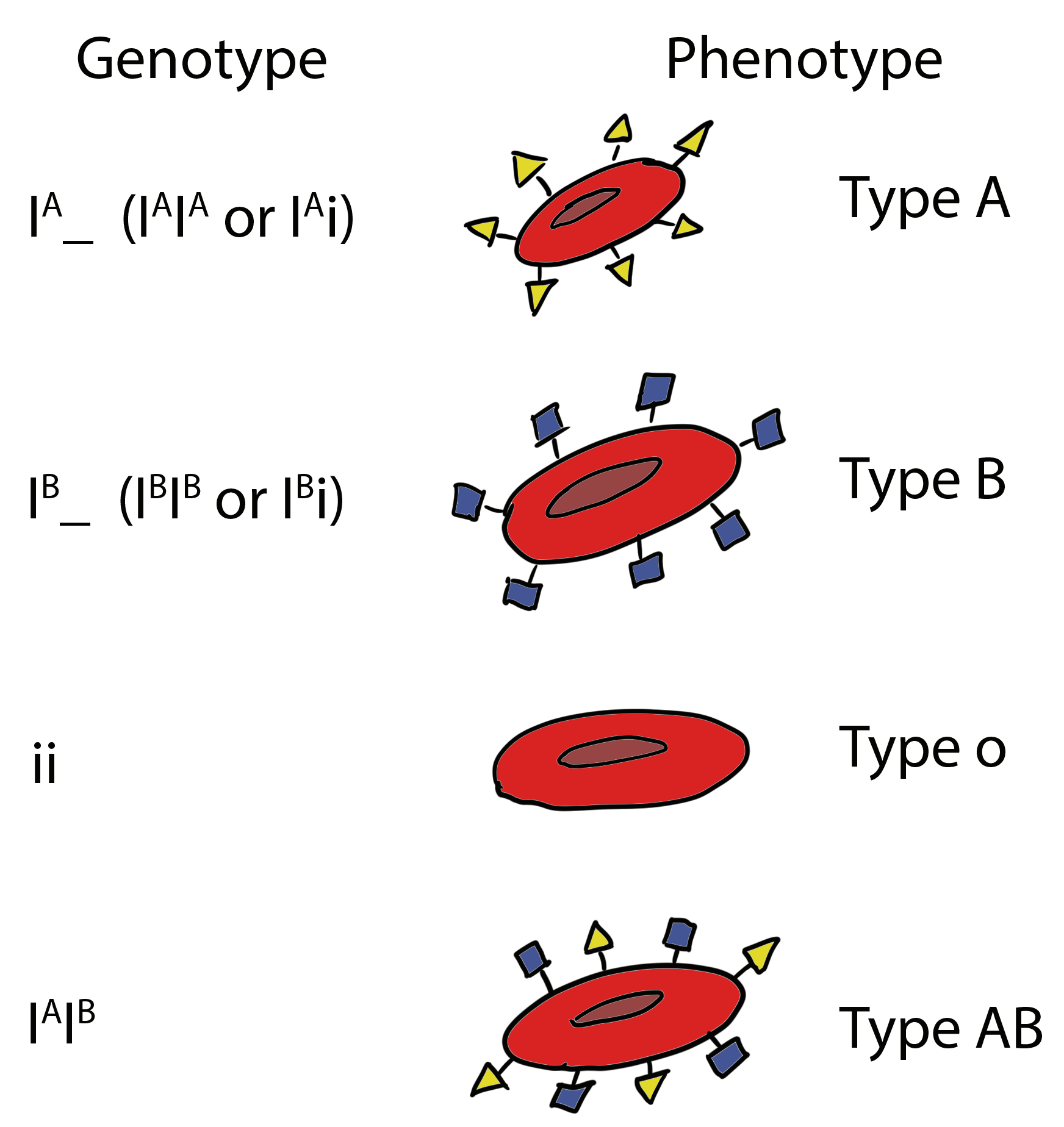
The ABO blood type is determined by three alleles that specify enzymes that turn the precursor molecule into a functioning antigen:
– IA (produces A antigen, antibodies against B in plasma);
– IB (B antigen, anti-A antibodies);
– iO (lacks either A or B antigens (no modification to the precursor molecule), carries both anti-A and anti-B antibodies).
IA and IB are co-dominant to each other and both are dominant to iO. Thus, the ABO blood group system is also an example of multiple alleles.
Epistasis, interactions between genes
Epistasis is a genetic state where the effects of a given gene on a biological trait are masked or enhanced by one or more other genes.
Epistasis refers to the observation that some phenotypes only arise when specific alleles of two (or more) genes interact with each other to cause a specific phenotype. Genetic interaction can proceed at many different levels. In most familiar cases, it is the genes’ products that interact with one another.
In humans, the Bombay phenotype is an example of epistasis. In the Bombay phenotype, the absence of a synthesis of a precursor antigen (H) can prevent the A or B antigens from being produced.
The H antigen is a precursor to the A and B antigens. If only recessive alleles for the H antigen are inherited (hh), the H antigen will not be produced. Subsequently, the A and B antigens also will not be produced. The result is an O phenotype by default since a lack of A and B antigens is the O type. In this case, an O blood type child can be born to parents with no iO allele.
Other examples of epistatic effects on traits include height and obesity.
Penetrance
Penetrance indicates the fraction of individuals of a particular genotype who show the expected phenotype.
An individual may have an abnormal genotype but show no phenotype.
Example: a human disease. If only 75% of AA individuals have the disease, the pene- trance is 75%.
Incomplete penetrance in humans: polydactyly (extra digits), an autosomal domi- nant condition.
Polydactyly doesn’t always express in all who carry the gene, but is passed on to the next generation even in those individuals who don’t express it (skips a generation). Polydactyly also shows epistasis.
Another example of penetrance: PKU (environmentally affected).
Other genes may “modify” the expected phenotype.
Expressivity
Expressivity refers to a variation in a phenotype in individuals carrying a particular genotype.
Example of varying expressivity in humans: Neurofibromatosis (NF).
NF is an autosomal dominant disease of the nervous system. Mild NF symptoms include various pigmented spots on the skin. Severe NF causes additional abnormalities (bone deformities etc.).
There are two types of NF. NF1 is due to a mutation in the neurofibromin gene located on chromosome 17 (17q11.2). This mutation can be caused by an insertion of a mobile genetic element (Alu). The frequency of NF1 in general population is 1 in 3,500.
NF2 is caused by a mutation in the gene that specifies neurofibromin 2, a tumor suppressor protein, located on chromosome 22 (22q12).
Example: Marfan Syndrome
Another example of variable expressivity is Marfan syndrome. Marfan syndrome is an autosomal dominant disease caused by a mutation in collagen formation. It affects about 1/5,000 live births. Symptoms of Marfan syndrome include skeletal, optical, and cardiovascular abnormalities. Skeletal abnormalities include arachnodactyly (long fingers and toes), extreme lengthening of the long bones, scoliosis, rib and sternum abnormalities, among others. Optical abnormalities almost always include ectopia lentis, a dislocation of the lens into the anterior chamber of the eye. Cardiovascular abnormalities may be numerous and include possible dissecting aneurysms, which are largely responsible for the shorter life span of Marfan syndrome patients as a group. Each patient may express all of the symptoms, or only a few. That is variable expressivity. Each patient with the mutant allele for Marfan syndrome expresses at least one of the symptoms, but the physician may have to look closely. Almost all are taller than average, but a lot of non-Marfan individuals are tall. Almost all have long fingers, but so do a lot of non-Marfan persons. Some may be very mildly affected and lead normal lives while others, more severely affected, have a shorter life expectancy. The disease is maintained in the population through recurrent mutations and the matings of less severely affected individuals with normal individuals. The extent of
severity of affected does not affect the severity of expression in the next generation, that is, the offspring of mildly affected individuals range from mildly affected to severely affected, with equal probability.
Famous people suspected to have had Marfan syndrome: Abraham Lincoln (1809-1865), Nicolò Paganini (1782-1840) (a famous Italian violinist, violist, guitarist, and composer).
Genetic mosaicism
In genetic medicine, a mosaic or mosaicism denotes the presence of two populations of cells with different genotypes in one individual, who has developed from a single fertilized egg.
Different types of mosaicism exist, such as gonadal mosaicism (restricted to the gametes) or tissue mosaicism; one of them is chimerism, where two or more genotypes arise from the fusion of more than one fertilized zygote in the early stages of embryonic development. In the more common mosaics, different genotypes arise from only a single fertilized egg cell, due to errors in early mitoses. In rare cases, intersex conditions can be caused by mosaicism where some cells in the body have XX and others XY chromosomes.
Cellular mosaicism in females most commonly results from a random inactivation of an X chromosome in females in early development, where all cells in an organism have the same genotype, but a different copy of the X chromosome is expressed in different cells. The coloring pattern of calico cats is the result of X inactivation. In an animal heterozygous for X-linked (associated with the X chromosome) alleles for fur color, different parts of the cat’s skin contain genes that produce differently colored fur, based on which alternative allele is expressed from one or the other X chromosome in the group of skin cell beneath a particular color patch.
Dosage compensation and the X-chromosome inactivation
Dosage compensation is a genetic regulatory mechanism, which operates to equalize the expression of characteristics determined by genes on the X chromosome so that they are equally expressed in the human XY male and the XX female.
Dosage compensation in humans proceeds via random inactivation of one of the two X chromosomes in female embryos. This inactivation happens by the 16-cell stage (days 3-4 post-fertilization in humans), when cells of the developing embryo are still in the totipotent state and before gene expression from X-linked genes takes place. At this stage, each embryonic cell randomly decides to inactivate either the paternally-derived X or the maternally-derived X. This “decision” is inherited by all of the descendants of the original cell. The inactivated X chromosome turns into a Barr body. Barr bodies can be seen in interphase nuclei as darkly staining heterochromatic masses.
There is an X-linked mutation that causes anhidrotic ectodermal displasia (absence of sweat glands). Females heterozygous for the mutation display large patches of skin lacking sweat glands. This is because the decision to inactivate the X is made randomly at early stages of development, leading to mitotic clones that eventually become patches of normal and mutant skin.
X chromosome inactivation is an example of epigenetic control of gene expression. Epigenetic control is excerted without changing the DNA sequence and usually involves modifications of chromatin packaging, making the DNA in certain regions of chromosomes, or even entire chromosomes, like in the case of X chromosome, inaccessible for processing. This means that genes in the “silenced” regions of chromosomes are not expressed. One way of “silencing” a chromosomal region is to methylate cytosines on the DNA. Methylation involves the attachment of methyl groups (-CH3), making the region containing methyl groups hydrophobic, which leads to the condensing of the chromatin in the methylated region, thus “hiding” it from being accessed by cellular proteins involved in gene expression. When located in a gene promoter, DNA methylation typically acts to repress gene transcription. DNA methylation is essential for normal development and is associated with a number of key processes including genomic imprinting, X-chromosome inactivation, repression of transposable elements, aging and carcinogenesis.
Genomic Imprinting
The pattern of inheritance when expression of a gene depends on whether it is inherited from the mother or the father is called genomic imprinting. Genomic imprinting is another example of epigenetic control and it involves DNA methylation.
The Prader-Willi syndrome (PWS) in humans is caused by the absence of part of the long arm (q11-13) of the paternally derived chromosome 15. In 70-80% of PWS cases, the region is missing due to a deletion. In normal individuals, certain genes from this region are not expressed from the maternal chromosome, but are expressed from the paternal chromosome. When these paternally derived genes are absent or disrupted, the PWS phenotype results. When this same segment is missing from the maternally derived chromosome 15, a completely different disease, Angelman syndrome, arises.
Additional challenges to the genotype-phenotype relationship
Polygenic Inheritance
Classic phenotypes correspond to one of two sorts: green v. yellow, wrinkled v. smooth. Such phenotypic distribution is usually attributed to traits under a single gene control. Traits specified by more than one gene express continuous variation. Continuous variation, also known as quantitative variation, is observed when a spectrum of phenotypes exist rather than two or three discrete, qualitative differences.
When variation is quantitative, phenotypes often follow a normal distribution, in which the extreme phenotypes are relatively rare, and intermediate phenotypes are the most common.
In polygenic inheritance, several genes contribute to the phenotype in an additive manner (i.e., enhancing or diminishing the trait’s expression, like height or the intensity of skin pigmentation).
If polygenic inheritance involves just a few loci with complete dominance, there may be several distinct phenotypes among the offspring.
Pleiotropy: one gene affecting many traits
The condition where a single gene has an effect on more than one phenotype is called pleiotropy. Examples of pleiotropy in humans include juvenile-onset diabetes mellitus and PKU.
PKU is an autosomal recessive disorder that affects 1 in 10,000 newborns in the US (1 in 50 is a carrier). PKU is one of the most common inborn errors in amino acid metabolism in humans. PKU results from mutations that least to the absence of phenylalanine hydroxylase (PAH) function. PAH converts phenylalanine to tyrosine. L-tyrosine is used to produce noradrenaline, adrenaline, dopamine. It is also a precursor of melanin. L-tyrosine aids in the functions of the adrenal, thyroid, and pituitary glands. Thyroxin (a thyroid hormone, made from L- tyrosine) is involved in regulation of metabolic rate, mental health, and growth rate.
Symptoms of PKU include cognitive disability, melanin deficiency (lighter skin, blue eyes) and related physiological manifestations. The most effective treatment is dietary restriction of phenylalanine from birth to adolescence.
The frequency of the PKU allele is higher than what would be expected for a recessive deleterious allele. Therefore, it has been suggested that there may be a heterozygous advantage in PKU that operates through protection against the toxic effects of ochratoxin A (Wolf, 1986). Ochratoxin A, an N-acyl derivative of phenylalanine, acts by competing with phenylalanine for phenylalanyl-tRNA synthetase, thus bringing protein synthesis to a halt. Doses of phenylalanine sufficient to raise the blood phenylalanine concentration reduce or reverse the toxicity of ochratoxin A.
This mycotoxin is produced by several species of Aspergillus and Penicillium that infest stored grains and other foods. The mild, wet climate of Ireland and West Scotland tends to encour- age the growth of molds. Furthermore, these areas have suffered repeated famines during which moldy food was eaten. Heterozygous women appear to have a lower spontaneous abortion rate (Eisensmith and Woo, 1992).
Another example of pleiotropy (in association with epistasis): red haired people may have a different perception of pain
The gene called MC1R causes 80% of all cases of red hair. (Other cases of red hair are due to mutations outside of the MC1R gene.) It is responsible for dark pigment deposition in hair. If an individual carries two recessive copies of the MC1R gene, the pigment is not deposited and the hair color appears red. Apparently, people who are homozygous for the inactive MC1R gene, have different pain sensitivity than the rest of us. Some red-haired people are more sensitive to pain, while others respond quite the opposite. The exact nature of this differential response to pain remains unknown, and the research on the genetic pathways of MC1R continues.
The Red Hair Gene
How often do you meet a person with red hair? It depends on where you are. In England and Scotland, for example, where the trait shows up at its highest frequency, the chance is about 1/30. Overall in the world, people with red hair comprise about 2% of the population.
The gene responsible for hair color is called MC1R. It is located on chromosome 16. The MC1R gene provides instructions for making a protein called the melanocortin 1 receptor. This receptor plays an important role in normal pigmentation. The receptor is primarily located on the surface of melanocytes, which are specialized cells that produce a pigment called melanin. Melanin is the substance that gives skin, hair, and eyes their color. Melanin is also found in the light-sensitive tissue at the back of the eye (the retina), where it plays a role in normal vision. The MC1R gene comes in several versions, called alleles.

The melanocortin 1 receptor controls which type of melanin is produced by melanocytes. When the receptor is activated, it triggers a series of chemical reactions inside melanocytes that stimulate these cells to make eumelanin (the brown and black melanin). If the receptor is not activated or is blocked, melanocytes make pheomelanin, which is responsible for lighter-colored pigmentation.
Common alleles of the MC1R gene are associated with normal differences in skin and hair color. Certain genetic variations are most common in people with red hair, fair skin, freckles, and an increased sensitivity to sun exposure.
These MC1R alleles lead to the production of the melanocortin 1 receptor with a reduced ability to stimulate eumelanin production, causing melanocytes to make mostly pheomelanin. One must inherit two copies of a particular red hair-associated allelic variant of the MC1R gene to actually have red hair. A person carrying a red hair-producing allele on one of the two chromosomes #16 (you inherit one from your mother and another from your father) and an allele producing a fully functional version of MC1R will have darker hair color. Those alleles that require just one copy to be present in the genome to phenotypically express the trait they specify are called dominant. The alternative alleles, like the red hair-causing MC1R allele, are called recessive.
Heritability, environmental effects and phenotypic plasticity
Heritability of a trait is defined as ratio of genetic variance to total phenotypic variance (genetic and environment): H2=V(G)/V(P). The parameter H2 is the broad-sense heritability and reflects all possible genetic contributions to a population’s phenotypic variance. Included are effects due to allelic variation (additive variance), dominance variation or which act epistatically (multigenic interactions), as well as maternal and paternal effects, where individuals are directly affected by their parents’ phenotype (such as with milk production in mammals). Pathological innumeracy, known as dyscalculia, is often associated with neurological lesions.
The genetic component of the phenotype’s variation of a particular trait (heritability) can be measured by comparing the trait’s expression in “affected” families with that of the general population. The genetic component has to show up more frequently in related individuals compared to the general population: twins > siblings > adopted children = general population.
How much of the phenotypic expression of a trait is inherited and how much of it is due to the environment? A meta-study of heritability correlations in twins published in 2015 by Nature Genetics (Polderman et al., 2015) revealed what geneticists have been suspecting all along: there’s an almost even split of the influence between environment and genetics on the expression of our phenotypic traits. The finding is based on a review of 2748 studies involving 14 million twin pairs from across 39 countries. The researchers were able to determine the contribution of genetics and the environment on the trait by measuring how similar various traits are between identical twins and non-identical twins. “If the trait is genetic then you would expect identical twins will be more similar than the non-identical twin. The more similar an identical twin to a non-identical twin then we can infer the trait is largely due to the genetic factor,” says Dr Beben Benyamin, on of the authors of the article from the Queensland Brain Institute in Australia. For all traits, the average heritability is about 0.49, or 49%, while the environment accounts for the remaining 51% of traits’ variation. However the heritability of different traits varies widely. For example, schizophrenia is 70% heritable, while the heritability of longevity in humans is well below 10% (Ruby et al., 2018). Height, one of the best-studied human traits with respect to heritability, is 86% heritable. At the same time, knowing the heritability of a trait does not provide information about which genes or environmental influences are involved, or how important they are in determining the trait.
Phenotypic plasticity and norm of reaction
Phenotype will depend upon genetic and environmental factors, and the interaction between them. Phenotypic plasticity is the ability of an organism to change its phenotype in response to changes in the environment. The range of phenotypic possibilities for a single genotype, as influenced by the environment, is called the norm of reaction. The term was introduced to genetics by W. L. Jahannsen, a Danish biologist who also contributed to the advancement of our understanding of the basic principles of selection through his studies on true-bred lines.
Key Takeaways
- Classical (Mendelian) genetics studies the relationship between the genotype and the phenotype.
- The Mendelian transmission genetics assumes a binary correspondence between a character and a gene that encodes that character.
- In reality, the relationship between the genotype and the phenotype is affected by a variety of factors.
- DNA methylation can affect gene expression in sex-dependent ways, based on which of the two parents contributes the affected chromosome.
A condition in which an organism contains one of each kind of allele.
Genotype is the particular combination of alleles for a particular gene or locus.
A grouping of genomic variants that tend to be inherited together.
Capable of giving rise to any cell type.
A tightly packed (and darker stained) form of chromatin usually associated with areas of low or no gene expression (such as centromeres and telomeres). Heterochromatic regions are heavily methylated.
