Chapter 10. Mutations
Mutations
A mutation is any change in the nucleotide sequence of a gene. A mutation occurs when genetic material is damaged or changed in such a way as to alter the genetic message carried by that gene. Mutations can be induced by a variety of factors called mutagens.
Mutations and their functional consequences
In terms of functional changes to gene product, mutations can be classified as loss of function and gain of function.
Loss of function mutations are usually recessive at the genetic level, because, most of the time, a functional product is still made by the undamaged copy of the gene (the wild type allele).
Gain of function mutations are usually dominant, because a new abnormally active product is made which may cause unregulated cellular changes.
A mutagen is an agent or substance that can bring about a permanent alteration to the physical composition of DNA such that the genetic message is changed. Once the gene has been damaged or changed the mRNA transcribed from that gene will now carry an altered message. The polypeptide made by translating the altered mRNA will now contain a different sequence of amino acids. The function of the protein made by folding this polypeptide will probably be changed or lost. Consequently, the phenotype of the organism carrying the mutation will be changed as well.
Mutations in the DNA of somatic cells may lead to a change in an individual phenotype, but will not be passed on to the next generation. Only mutations in reproductive cells are inherited.
Types of mutagens:
- Spontaneous (unrepaired errors in DNA replication).
- Chemical (caused by a chemical agent, such as benzine or hormones (estrogen, testosterone)).
- Physical (caused by a physical agent, such as radiation).
- Biological (caused by transposable elements and viruses).
Environmental Factors: Radiation
Radiation is a process by which energy travels through space. Exposure to radiation is unavoidable and comes from a wide variety of sources both natural occurring and due to human activity.
The Chornobyl Nuclear Accident
On April 26, 1986, a reactor at the Chornobyl nuclear power plant in Ukraine experienced a catastrophic failure, which led to a release of vast amounts of radiation into the atmosphere. Radiation traveled around the world and within 24 hours was reported as far away as Canada. The bulk of radioactive material fell on the 20-km territory in the immediate vicinity of the plant. This “exclusion zone” became permanently unsuitable for human habitation. People traveling to the zone have to wear protective gear patterned after an astronaut suit, since it is as dangerous for a human body to be in the zone as being in an outer space. The most prominent health result of exposure to Chornobyl radiation is a 1,000-fold increase in thyroid cancer rates in children and adolescents who were exposed to radiation effects at the time of the accident. Elevated rates of hydrocephaly (see picture), leukemia, and various birth defects have been reported in the affected population. Cancer rates are projected to increase in the larger population in the affected territory for another 20 years.

Point mutations
Point mutations are single base (nuceotide) changes (additions, deletions, substitutions) in a sequence of DNA that specifies a cellular product.
Base substitutions are classified as follows.
A nonsense mutation creates a stop triplet where none previously existed.
A missense mutation changes the triplet code of one amino acid for a different one. A silent mutation has no effect on the code.
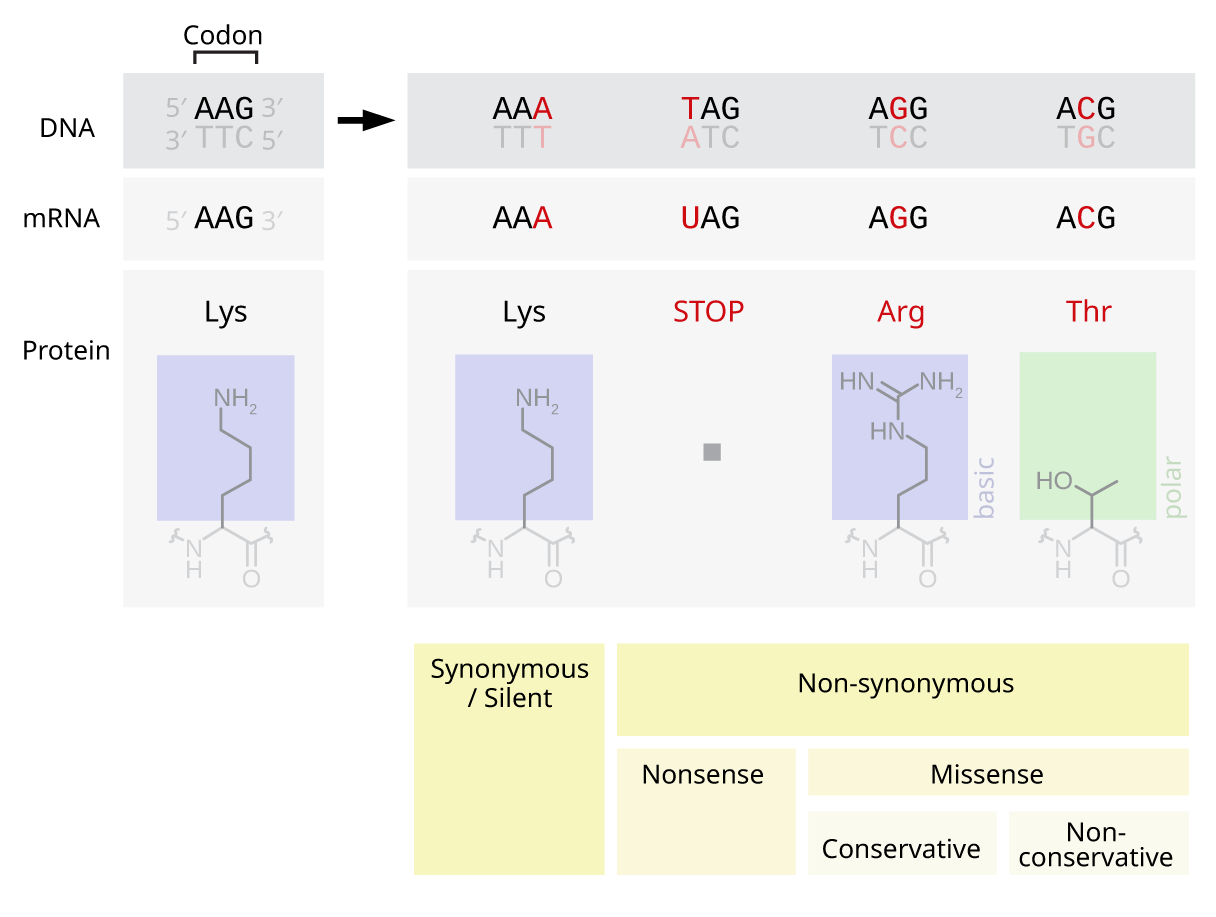
Within a gene, deletions or insertions of one or two nucleotides result in a frame shift. Larger deletions or insertions (≥2 nucleotides) may result in cessation of transcriptional activity in that gene.
Mutations with Expanded Trinucleotide Repeats
Trinucleotide repeats are a class of mutations associated with a number of genetic disorders. Trinucleotide repeats can expand in successive generation, leading to an increase in the size of genes in which these repeats are found and, in some cases, lead to an inherited genetic disorder. Disease severity may be related to the size of the expanded gene. The expansion of nucleotide triplets causes the type of inheritance called allelic expansion.
Allegan County Sheep Could Hold Key To Treating Huntington’s Disease
Joe LaFurgey and 24 Hour News 8 web staff
Published: May 6, 2016, 6:34 pm
https://www.woodtv.com/news/allegan-county-sheep-could-hold-key-to-treating-huntingtons-disease/
The reposting of the article adheres to the Fair Use application of 17 U.S. Code § 107
HOPKINS, Mich. (WOOD) – Research coming out of an Allegan County farm could bring hope to the 30,000 people who suffer from Huntington’s disease and the 200,000 people at risk of getting it. Mike Ludlam’s sister, Dori was part of that statistic. He lost her to Huntington’s disease four years ago.
“You basically just watch somebody deteriorate in front of your eyes. [They] go from such a strong person… [to] being helpless,” said Ludlam, who owns Windswept Farms near Hopkins.
Huntington’s disease is a genetic disorder that damages brain cells and patients’ mental and physical abilities, eventually taking their life. But an article Ludlam recently came across could be a game-changer.
The report described a genetic disorder in some sheep which cause the animals to overproduce the GM1 ganglioside, which is a substance that helps protect brain cells. It’s also a substance lacking in people with Huntington’s disease. Suddenly, the Ludlams’ decision years ago to raise sheep took on new meaning. “We have sheep. We’re sheep farmers. This is a no brainer,” said Heather Ludlam, who is also a veterinarian. The couple helped jumpstart research involving breeding sheep with the genetic disorder that produces the additional GM1 and extracting the substance for use in treating Huntington’s.
GM 1 isn’t a cure, but it could bring an effective treatment for the disease. The early results are promising and piqued the interest of U.S. Rep. Fred Upton, who toured the Ludlam’s farm Friday. “Do they have a time line of when they finish that first study?” Upton asked Heather Ludlam. “They’re hoping, I’d say, between two and three years. It could come down to funding,” she answered.
Upton is the primary sponsor of the 21st Century Cures Act. The act would provide additional funding for the National Institutes of Health, as well as streamline the process of getting treatments to patients quicker.
For Mike Ludlam, Upton’s visit was one more step in the process. “We’ve kind of figured with this project, we don’t have anything to lose,” Mike Ludlam dded. “We’re just going to keep knocking on doors and trying to get the story out… to get it in front of that one person.”
The Position of the Mutation in a Gene Affects the Phenotype
Cystic fibrosis (CF) is an autosomal recessive genetic disorder affecting the lungs, pancreas, liver, intestine, and reproductive tract. It is characterized by abnormal transport of chloride ions across epithelium, leading to thick, viscous secretions.
The cystic fibrosis transmembrane conductance regulator (CFTR) gene is located on chromosome 7, position 7q31. A defect in the CFTR gene leads to clogging of bronchial tubes with thick mucus.
In normal cells, the CFTR protein acts as a channel that transports chloride into and out of cells, and regulates the passage of salts. It is made up of five domains: two membrane-spanning domains that form the chloride ion channel, two nucleotide- binding domains (NBDs) that bind and hydrolyze ATP and a regulatory domain.
There are currently around 1400 mutations known to be associated with CF. The most common mutation that leads to the CF disease is a P508 deletion (delta508). The removal of phenylalanine 508 from the N-terminal region of the NBD1 leads to the disruption of the proper folding of the N-terminal, leading to its inability to bind ATP. Thus, the P508 deletion affects the function of the entire CFTR protein.
The P508 mutation is considered to be associated with overdominance. Studies in mice and human cell cultures show that cells homozygous for the delta508 mutation make cells impermeable to Salmonella typhi, the bacterium that causes typhoid fever in humans. The protective effect of delta508 against S. typhi manifests even when only one delta508 allele is present. Gastrointestinal submucosa cells of the heterozygous for delta508 mice reduce the translocation ability of S. typhi by 86% (Pier et al., 1998).
Biological Mutagens: Transposable elements
Transposable genetic elements (TEs, also called transposons or mobile genetic elements) comprise a diverse family of DNA sequences that are capable of moving to new sites in genomes either directly by a cut-and-paste mechanism (transposons) or indirectly via an RNA intermediate (retrotransposons). Transposons were first discovered by a Ukrainian geneticist Serhij Hershenson in the 1920s in Drosophila (Hershenson, 1928, 1939). The worldwide scientific community did not appreciate this discovery for nearly two decades, until TEs were rediscovered in maize plants by the American geneticist Barbara McClintock in the mid-1940s. For several decades, transposons were considered something of a genetic oddity. In the 1980s, TEs were labeled as “junk” DNA, along with other non-coding sequences in the human genome. In the 1980s, the biological importance of TEs started to come into focus.
Transposable elements are found in genomes of all living things. Transposons are the single largest component of the genetic material of most eukaryotes. Mobile genetic elements constitute more than half of the DNA in most higher eukaryotes.
There are two types of TEs:
- Class II transposons. These consist of DNA that moves directly from place to place (cut-and-paste).
- Class I transposons. These are retrotransposons that first transcribe the DNA into RNA and then use reverse transcriptase to make a DNA copy of the RNA to insert in a new location (copy-and-paste).
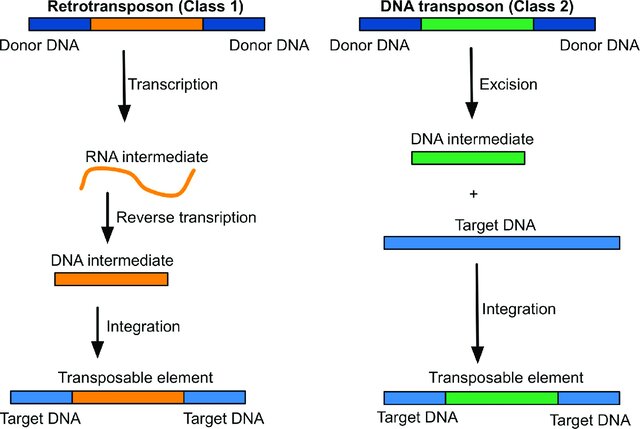
Human retrotransposons include autonomous (can move on their own) Long Interspersed Elements – LINEs (representing 17% of the genome), and non-autonomous Alu elements (11% of human genome). LINEs and Alu elements are the only TEs in the human genome that are still active.
Sequences derived from TEs are found within introns of most mammalian genes. The proliferation and evolution of retrotransposons have had multiple impacts on organismal genomes. Retrotransposition can destabilize the genome, shaping genomic landscapes by insertional mutagenesis, deletions and gene rearrangements (see Chapter 16).
Transpositional activity of DNA (class II) TEs is predominantly deleterious and mostly restricted in its effect to the somatic genome (Nikitin & Shmookler Reis, 1997). Transpositional activity of RNA (class I) TEs (retrotransposons) can have major evolutionary consequences, exerted through facilitation of ectopic recombination and changes in gene expression.
In humans, Alu sequences are a major factor contributing to non-allelic homologous recombination and insertional mutagenesis. The table below provides examples of insertional mutagenesis cased by Alu sequences linked to human diseases.
| Gene | Disorder | Element | Mechanism | Reference |
| NF1 | Neurofibromatosis | AluYa5 | Intron insertion/frameshift | Wallace et al. 1991 |
| ChE | Acholinesterasemia | AluYb8 | Exon insertion | Muratani et al. 1991 |
| F9 | Hemophilia B | AluYa5 | Exon insertion | Janicic et al. 1995 |
| CASR | Familial hypocalciuric hypercalcemia | AluYa4 | Exon insertion | Bai et al. 1997 |
| ADD1 | Huntington disease | Alu | Intron | Hutchinson et al. 1993 |
Insertions of mobile genetic elements or transposons (see Chapter 1) contribute to disease by either disrupting a coding region or a splice signal. Although insertions of such human transposons as Alu causing disease are broadly spread throughout the genome, some genes seem more prone to disease-causing insertions of this type. Fourteen new Alu insertions inactivating the NF1 gene have been reported, representing 0.4% of known mutations in this gene. Similarly, many diseases caused by non-allelic homologous recombination between Alu elements have been reported as well. Although these events are also broadly spread throughout the genome, some regions, such as the MSH2, VHL and BRCA1 genes, are much more subject to this instability than others. Most Alu-related genomic instability events will either have no major functional consequence, and over many generations simply be lost from the human population gene pool through random fixation, or be deleterious and therefore lost through negative selection. Thus, the events described above represent only a tiny proportion of the overall genetic instability in the human population caused by such elements.
Key Takeaways
- Any alteration of a DNA sequence is characterized as a mutation.
- Point mutations, a change in one or two nucleotides on DNA strand, are caused by mutagenes and are classified based on their functional consequences.
- Transposable elements are a major contributor to mutagenesis in humans.
