Chapter 16. Human Evolutionary Genetics

Evolution: From Darwin to Modern Synthesis
The union of the evolutionary theory by natural selection of Darwin and Wallace with subsequent discoveries in classical and molecular genetics is termed the Modern Evolutionary Synthesis.
The seminal work on modern evolutionary synthesis was published in 1937 by Theodosius Dobzhansky (Feodosii Dobrzhanskyi in Ukrainian, 1900-1975), a Ukrainian-American geneticist and evolutionary biologist whose work had a major influence on the 20th-century thought and research on genetics and evolutionary theory.

Dobzhansky was a pivotal figure in the development of the modern evolutionary synthesis. Dobzhansky’s most influential work, *Genetics and the Origin of Species* (1937), demonstrated how genetic mutations and variations within populations could lead to evolutionary changes over time. This book played a crucial role in bridging the gap between genetics and evolutionary theory, showing that mutation, genetic recombination, and natural selection could drive the evolution of species.
Dobzhansky conducted extensive genetic experiments with Drosophila fruit flies. His work provided empirical evidence for the role of genetic variation in natural populations and how these variations could lead to adaptive changes. He showed that genetic diversity within populations is crucial for the process of natural selection to operate. Alongside natural selection, Dobzhansky highlighted the importance of genetic drift—random changes in gene frequencies in small populations. He demonstrated that genetic drift could significantly affect the genetic makeup of populations, especially in isolated groups.
Dobzhansky also made significant contributions to the understanding of speciation, the process by which new species arise. He emphasized the role of reproductive isolation—mechanisms that prevent different species from interbreeding—in maintaining species boundaries and promoting the emergence of new species. He advocated for “population thinking,” the idea that evolutionary change occurs within populations rather than individuals. This perspective was essential for understanding the genetic structure of populations and how it influences evolutionary dynamics.
Dobzhansky’s contributions to the modern evolutionary synthesis were foundational. His integration of genetics with evolutionary theory, experimental evidence, and comprehensive approach to studying population genetics and speciation significantly advanced the understanding of how evolutionary processes shape the diversity of life.
In his “Genetics and the Origin of Species” Dobzhansky defined evolution as “a change in the frequency of an allele within a gene pool”. Dobzhansky’s work established that mutations are the source of genetic variation from which natural selection chooses the variants that confer better survival and reproduction.
Molecular clock
Generally stated, molecular clock is the average rate at which a genome accumulates mutations. Since its proposal in the 1960s, the molecular clock has become an essential tool in many areas of biology, including systematics, molecular ecology, and evolutionary genetics. The molecular clock hypothesis posits that DNA and protein sequences evolve at a relatively constant rate over time and among different organisms. This constancy implies that the genetic differences between any two species (or any two DNA sequences) are proportional to the time since they last shared a common ancestor. Consequently, molecular clock serves as a highly valuable method for estimating evolutionary timescales, both within and between species.
The molecular clock hypothesis was originally proposed by Emile Zuckerkandl and Linus Pauling based on empirical observations. It gained theoretical support when biologist Motoo Kimura developed the neutral theory of molecular evolution in 1968. Kimura suggested that many new mutations do not affect evolutionary fitness, so natural selection neither favors nor disfavors them. Over time, these neutral mutations would either become fixed in a population or be lost entirely through genetic drift. Kimura demonstrated that the rate at which neutral mutations become fixed (the substitution rate) is equivalent to the rate at which new mutations appear (the mutation rate). If the mutation rate is consistent across species, the substitution rate would remain constant throughout the tree of life.
Subsequent research has shown that rates of molecular evolution can vary significantly among organisms. Nevertheless, the molecular clock remains a valuable tool in evolutionary studies. Researchers have refined the concept by developing “relaxed” molecular clocks, which allow the molecular rate to vary among lineages within limits. There are two major types of relaxed-clock models. The first assumes that the rate varies over time and among organisms, but this variation occurs around an average value. The second allows the evolutionary rate to “evolve” over time, assuming it is tied to other evolving biological characteristics. For instance, there is evidence that substitution rates are influenced by an organism’s metabolic rate.
Calibration is crucial when using either strict or relaxed molecular clocks. For example, if researchers have two DNA sequences differing by 5%, they cannot determine the divergence time without knowing the rate of divergence. Calibration requires an absolute age for an evolutionary divergence event, such as the split between mammals and birds. This can be estimated from the fossil record or by correlating divergence with geological events of known age, like the formation of a mountain range that initiated speciation. Once the evolutionary rate is calculated using such calibration, it can be applied to other organisms to estimate the timing of evolutionary events. When applied correctly, the molecular clock can provide enlightening date estimates for evolutionary events that are difficult to study through the fossil record alone. Relaxed-clock methods help account for rate variation, offering insights into the biological processes influencing the molecular clock’s pace.
Genome rearrangements and evolution
As many as 99% of human genes are conserved across all mammals. The functionality of many genes is virtually the same among many organisms. It is highly unlikely that the same gene with the same function would spontaneously develop among all currently living species.
Humans and mice have similar genomes, but their genes are ordered differently. A total of about 245 chromosomal rearrangements separate human and mouse genome. Among these, there are reversals, fusions, fissions, and translocations.
Significant chromosomal rearranging occurred between the diverging point of humans and mice. For example, human chromosome 3 contains homologous sequences to at least five mouse chromosomes. The homology between chromosome regions of different species is called synteny.
Human and mice chromosome homology
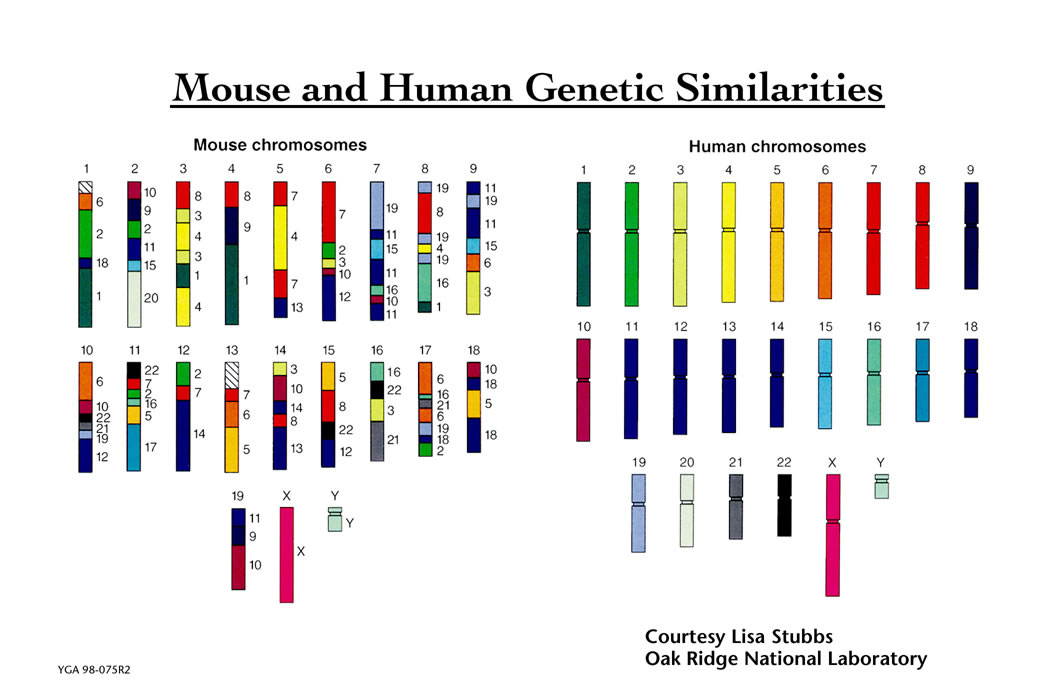
Credit: Lawrence Livermore National Laboratory. The use of the image adheres to the Fair Use application of 17 U.S. Code § 107.
Role of Transposable Elements in Evolution
Some TEs contain DNA binding site motifs for transcription factors and have rewired the transcriptional regulatory networks in which these transcription factors function by introducing many new binding sites via their insertions throughout the genome. TE-derived sequences also appear in RNA transcripts where they can interact with RNA binding proteins (RBPs), which also often have preferred binding site motifs. In perhaps the most understood and interesting example, the antisense strand of Alu elements contains motifs that recruit the cell’s splicing machinery and have thus introduced hundreds of novel exons into various protein coding genes. Sequence derived from TEs has also been implicated in both degradation and increasing the translation of RNA transcripts.
Chromosomal Differences Between Humans and the Great Apes
Humans share a branch of mammalian phylogenetic tree with the great apes of the Old World (chimpanzee, gorilla, and orangutan). Collectively, humans, the great apes, and their ancestors are called hominids. From the genetics standpoint, humans and the great apes are very similar in all but one major aspect. All great apes have 48 chromosomes (n=24), while humans have 46 (n=23). This karyotypic difference was caused by the fusion of two ancestral chromosomes in the ancestor of the hominin lineage to form human chromosome 2 and subsequent inactivation of one of the two original centromeres. The great apes retained the two ancestral chromosomes (2A and 2B).
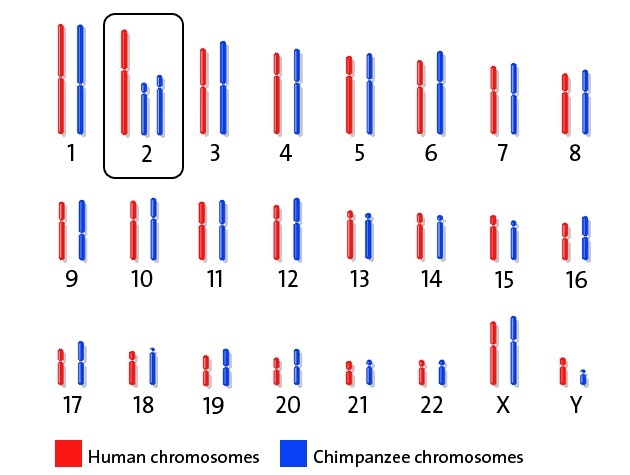
As a result of this fusion, sequences that once resided near the ends of the ancestral chromosomes are now located in the middle of chromosome 2, near the borders of bands 2q13 and 2q14.1.
Human evolution continues: 2n=44?
Reciprocal translocations are an exchange of material between non-homologous chromosomes (1/625 newborns). In Robertsonian translocations, long arms of acrocentric chromosomes fuse near the centromere region with loss of the short arms.
Robertsonian translocations, such as 14q/21q, can be inherited. A 2n=45, 14q/21q individual would be fertile and would produce gametes with n=23 and n=22; 14q/21q.
Suppose a 45-chomosome human meets a mate with the same translocation and the same karyotype. If they were to have children, there would be a 50% chance for their offspring to have a 2n=44 karyotype.
Such a scenario is not far fetched. There have been documented cases of fertile adult individuals with a 44-chromosome karyotype. A recent such case from China was described in 2013 (Wang et al., 2013). It describes a Robertsonian translocation family, one of them who came from a consanguineous marriage has the previously undescribed balanced human karyotype 44,XY,der(14;15)(q10;q10), der(14;15)(q10;q10). This man had fewer chromosomes but is actually missing very few genes. Instead, he has two chromosomes stuck to two other chromosomes. More specifically, both his chromosome 14’s are stuck to his chromosome 15’s. Thus, he is missing chromosome 15 pair and his chromosome 14 pair is a fusion chromosome. This is essentially what happened in the lineage leading to humans when we departed from the great apes due to a similar chromosomal fusion and became our own distinct species.
Human Origins

The human family tree spans over 7 million years, when our ancestors split from the ancestors of modern chimpanzees. There are different scenarios proposed by human evolutionary biologists, anthropologists and paleontologists over the last 100 years regarding Homo sapiens evolution. There is no single scenario exists at the present time that satisfies everybody. It became even more contentious in the last two decades with the advent of ancient DNA (aDNA) analysis. One of the best supported by scientific evidence scenarios is presented here.
The earliest hominin fossils come from Bulgaria and Greece and belong to the genus Graecopithecus that lived during the late Miocene 7.2 million years ago (MYA) (Fuss et al., 2017). The emergence of the genus Homo is currently dated to about 3.3 MYA (Püschel at al., 2021) , overlapping with the latest surviving members of the genus Australopithecus, proposed to have evolved in Africa some 4.4 MYA. Australopithecines, in turn, likely descended from an early African hominin genus called Ardipithecus, which origin is dated to around 6 MYA. Significant gaps still remain in our understanding of the early hominin evolution. It is even unclear whether any of the above-mentioned hominins are in the direct line of modern human descent.
The clear human ancestor, Homo erectus, emerged around 2.1 MYA. The Erectus fossils have been found in Africa, but most of the fossil evidence of the Erectus hominins comes from outside of Africa, from the Caucasus Mountains, China and Southeast Asia. The emergence of H. erectus followed what is, arguably, one of the most important genetic events in the ancestral human genome.
About 2.4 MYA a mutation in the Myosin Heavy Chain gene, MYH16, took place in the ancestor of modern humans. This frameshift mutation led to the inactivation of the the predominant myosin heavy chain, responsible for the powerful muscles supporting the mandibles of non-human hominids. The loss of this protein isoform led to significant size reductions in individual muscle fibers and entire masticatory muscles in the hominin lineage. These muscles connect at the saggital crest, a bony ridge at the top of the skull of hominids. The pressure that masticatory muscles exert on the skull constricts the size of the cranial vault. The relaxing of this constriction due to the MYH16 mutation led to the shift of the bulk of the support of the chewing function to the products of the MYH1, –2 and –7 genes in in hominins, the temporalis muscles that have a lower attachment point on the cranium (the temporal fossa). Such removal of the cranial pressure by the masticatory muscles may have led to the potentially unrestricted growth of the cranial capacity, resulting in a rapid increase in the brain size and complexity (encephalization) of the members of the Homo genus. This change is evident anatomically in the remains of H. erectus, which display a relatively gracile masticatory apparatus starting 2 MYA (Stedman et al., 2004).
The MYH16 mutation appeared at a time of major environmental transformations. A major climatic shift was taking place at the onset of Quaternary Glaciation ca. 2.58 MYA, changing the landscape in east Africa from woodland to open grasslands. The ecological niche change combined with the MYH16 mutation conceivably led to an in situ speciation event, producing the Homo lineage.
Homo sapiens emerged as a species around 300 thousand years ago (KYA). Currently the oldest remains of early modern humans come from the site of Jebel Irhoud in Morocco and are dated to 286 ± 32 KYA (Hublin et al., 2017). The Anatomically modern human (AMH) remains with essentially the same cranial morphology as the humans living today come from Greece (Apidima Cave, 210 KYA), Israel (Mount Caramel, 177 KYA, Skhul and Qafzeh caves, 125 to 90 KYA), and China (Daoxian and Zhirendong, between 120 and 80 KYA).
Evidence from mitochondrial DNA (mtDNA) suggests an AMH dispersal out of Africa ca. 130 KYA reached southeast Asia, subsequently moving back to Africa ca. 70 KYA, and dispersing out of Africa again ca. 60-50 KYA (Cabrera et al., 2018). It is likely (though not well documented at the moment) that these “pendular” migrations in and out of Africa have been taking place continuously throughout the history of the H. sapiens species, and that the most “successful” dispersal episode lead to a permanent colonization of the planet by H. sapiens, leaving a continuous genetic trail outside of Africa starting ca. 60-50 KYA.
Paleontological evidence suggests that there were several lineages of Homo co-existing in various parts of the world ca. 700-30 KYA. In Africa, H. erectus (2.1 MYA-120 KYA) coexisted with H. sapiens idaltu, an extinct Sapiens lineage (160 KYA), the Omo River hominins (possibly H. sapiens, 195 KYA), and Jebel Irhoud H. sapiens (286 KYA). In Asia, H. erectus sensu lato co-existed with several distinct Homo lineages and Erectus subspecies, including H. floresiensis on the island of Flores in Indonesia (700-12 KYA), H. luzonensis, a dwarf lineage of H. erectus from Callao cave in the Philippines (67 KYA), H. erectus soloensis from the Solo river in Java, Indonesia (550-35 KYA), Homo sp. altai (Denisovans, 100-30 KYA, H. neanderthalensis altai (300-70 KYA), as well as Homo lineage hybrids. In Europe, H. sapiens overlapped with Neanderthals territorially and even culturally, until about 40 KYA.
There is ample genetic evidence suggesting that AMHs have interbred, on a number of occasions, with our closest cousins, Neanderthals (Homo sapiens neanderthalensis) and Denisovans (Homo sp. Altai). We know about the latter from just a phalanx and three molars, all found in the same cave, called Denisova, in the Altai Mountains (southwestern Siberia). Archaeogeneticists (scientists who work with ancient DNA) were able to extract DNA out of these remains and reconstruct the Denisovan genome.
Based on analyses of their nuclear genome, Denisovans are a sister group of Neanderthals, although the mtDNAs of Neanderthals and present-day humans share an mtDNA ancestor more recently with each other than with Denisovans.

Based on the studies of the genomes of modern humans, Denisovans and Neanderthals, it is becoming apparent that modern humans carry a small percentage of Neanderthal and Denisovan genomes and that, collectively, modern humans had retained almost 50% Neanderthal DNA. Interestingly, the percentage of Denisovan DNA in modern humans is higher in southeast Asia and Austronesia, while modern Europeans harbor more Neanderthal ancestry.
Nose shape gene inherited from Neanderthals
by University College London
MAY 8, 2023
The repost of the article adheres to the Fair Use application of 17 U.S. Code § 107.
Humans inherited genetic material from Neanderthals that affects the shape of our noses, finds a new study led by UCL researchers.
The new Communications Biology study finds that a particular gene, which leads to a taller nose (from top to bottom), may have been the product of natural selection as ancient humans adapted to colder climates after leaving Africa.
Co-corresponding author Dr. Kaustubh Adhikari (UCL Genetics, Evolution & Environment and The Open University) said, “In the last 15 years, since the Neanderthal genome has been sequenced, we have been able to learn that our own ancestors apparently interbred with Neanderthals, leaving us with little bits of their DNA.
“Here, we find that some DNA inherited from Neanderthals influences the shape of our faces. This could have been helpful to our ancestors, as it has been passed down for thousands of generations.”
The study used data from more than 6,000 volunteers across Latin America, of mixed European, Native American and African ancestry, who are part of the UCL-led CANDELA study, which recruited from Brazil, Colombia, Chile, Mexico and Peru. The researchers compared genetic information from the participants to photographs of their faces—specifically looking at distances between points on their faces, such as the tip of the nose or the edge of the lips—to see how different facial traits were associated with the presence of different genetic markers.
The researchers newly identified 33 genome regions associated with face shape, 26 of which they were able to replicate in comparisons with data from other ethnicities using participants in east Asia, Europe, or Africa.
In one genome region in particular, called ATF3, the researchers found that many people in their study with Native American ancestry (as well as others with east Asian ancestry from another cohort) had genetic material in this gene that was inherited from the Neanderthals, contributing to increased nasal height. They also found that this gene region has signs of natural selection, suggesting that it conferred an advantage for those carrying the genetic material.
First author Dr. Qing Li (Fudan University) said, “It has long been speculated that the shape of our noses is determined by natural selection; as our noses can help us to regulate the temperature and humidity of the air we breathe in, different shaped noses may be better suited to different climates that our ancestors lived in. The gene we have identified here may have been inherited from Neanderthals to help humans adapt to colder climates as our ancestors moved out of Africa.”
Co-corresponding author Professor Andres Ruiz-Linares (Fudan University, UCL Genetics, Evolution & Environment, and Aix-Marseille University) added, “Most genetic studies of human diversity have investigated the genes of Europeans; our study’s diverse sample of Latin American participants broadens the reach of genetic study findings, helping us to better understand the genetics of all humans.”
The finding is the second discovery of DNA from archaic humans, distinct from Homo sapiens, affecting our face shape. The same team discovered in a 2021 paper that a gene influencing lip shape was inherited from the ancient Denisovans.
The study involved researchers based in the UK, China, France, Argentina, Chile, Peru, Colombia, Mexico, Germany, and Brazil.
More information: Automatic landmarking identifies new loci associated with face morphology and implicates Neanderthal introgression in human nasal shape, Communications Biology (2023). DOI: 10.1038/s42003-023-04838-7
Based on the mtDNA divergence the intermixing between Neanderthals and H. sapiens that can still be traced today in modern human genomes likely took place outside of Africa as early as 250 KYA, but no later than 110 KYA (Kuhlwilmet al., 2017). However, there were multiple admixtures between the two hominins the descendants of which did not survive to the present day.
The Oase 1 hominin from Romania, 39.5±2.5 KYA, who derived 4.8-7.3% of genome from Neanderthals, had a Neanderthal ancestor 4-6 generations back, with largest Neanderthal-derived chromosomal segments located on chromosomes 4, 5, 6, 9, 12. The Ust’-Ishim individual from west Siberia, 45 KYA, had 2.3% of his genome coming from Neanderthals, with the latest Neanderthal ancestor 13-7 KYA back. His chromosome 21 was largely Neanderthal-derived.
However, current genetic evidence suggests that in the last five thousand years of Neanderthal and modern human overlap in Eurasia, there was little, if any, biological interaction between the two groups. Such as, the Late Neanderthals (47-39 KYA) from west Europe and the Caucasus show no genetic introgression from modern humans.
“Thus we have given to man a pedigree of prodigious length, but not, it may be said, of noble quality. The world, it has often been remarked, appears as if it had long been preparing for the advent of man: and this, in one sense is strictly true, for he owes his birth to a long line of progenitors. If any single link in this chain had never existed, man would not have been exactly what he now is. Unless we wilfully close our eyes, we may, with our present knowledge, approximately recognise our parentage; nor need we feel ashamed of it. The most humble organism is something much higher than the inorganic dust under our feet; and no one with an unbiassed mind can study any living creature, however humble, without being struck with enthusiasm at its marvellous structure and properties.”
Charles Darwin, “The Descent of Man, and Selection in Relation to Sex”, 1889, p. 165
Key Takeaways
- Recent data indicate that ca. 70KYA Eurasia was populated by archaic hominins such as Neanderthals, Denisovans, their hybrids, as well as descendants of H. Habilis and H. erectus in southeast Asia, plus transient Anatomically Modern Humans (AMHs).
- The bulk of the interbreeding between resident archaic hominins and transient AMHs took place prior to 150KYA.
- The latter transmitted admixed traits to the most recent African migrants of the most recent major dispersal throughout Eurasia ca. 60-50KYA.
- Biological contacts between archaic hominins and AMHs that left genetic traces in the latter were sporadic after 150KYA and ceased altogether after ca. 45 KYA.
- Reasons: separation of ecological niches and subsequent speciation.
- Archaic hominin contribution to modern genomes:
- Neanderthals to Eurasians: 1-4%
- Neanderthals to Africans: none directly; 0.3% from recent admixture with modern Eurasians.
- Collectively, 50% of Neanderthal DNA can be retrieved from modern humans.
- 3–5% of the DNA of Melanesians and Aboriginal Australians and around 7%-8% in Papuans deriving from Denisovans.
The homology between chromosome regions of different species.
The taxonomical division that includes all modern and extinct great apes, modern and extinct hominis, and all their ancestors.
The taxonomical group consisting of anatomically modern humans (Homo sapiens), extinct human species and all our immediate ancestors (including members of the genera Homo, Australopithecus, Paranthropus and Ardipithecus).
